RESEARCH PROJECTS & FUNDING
The Nwosu Lab is pursuing research projects around four main areas, namely, disrupting cellular use of glutathione (GSH), targeting nutrient transporters, overcoming drug resistance/finding new therapeutic combinations or strategies, and characterization of genes that are potentially relevant therapeutic targets or biomarker candidates. These research interests are interconnected but may also be pushed in different interdisciplinary directions so long as the focus remains on solving basic or disease-related metabolism problems and or cancer therapeutic challenges. The advantage is that trainees have the opportunity to contribute to each other’s work, thereby increase their chances of publication. Prospective undergraduates, graduate/rotation students or postdocs are encouraged, but are not required, to express interest in one or more of the projects below when contacting Dr. Nwosu. If you are interested in the lab but not sure of which project to work on still reach out.
Disrupting GSH dependency
We have data indicating that some pancreatic cancer cells highly depend on cysteine, which they use to produce glutathione (GSH). GSH neutralizes oxidative stress in cells. The ability of cancer cells to generate and use GSH is one protective mechanism through which they resist stress, survive and grow. We have also found that genes encoding certain enzymes responsible for GSH metabolism are highly expressed in pancreatic cancer. These observations suggest that GSH and pathways involved in its metabolism are important in pancreatic cancer (Nwosu et al., unpublished). The Nwosu Lab is focused on various questions around how pancreatic cancer access and use GSH, which enzymes or regulators are the most important targets for preventing GSH usage, and possible opportunities for treating human pancreatic cancer by disrupting GSH utilization (Figure 1). This project is currently the main focus of the lab.

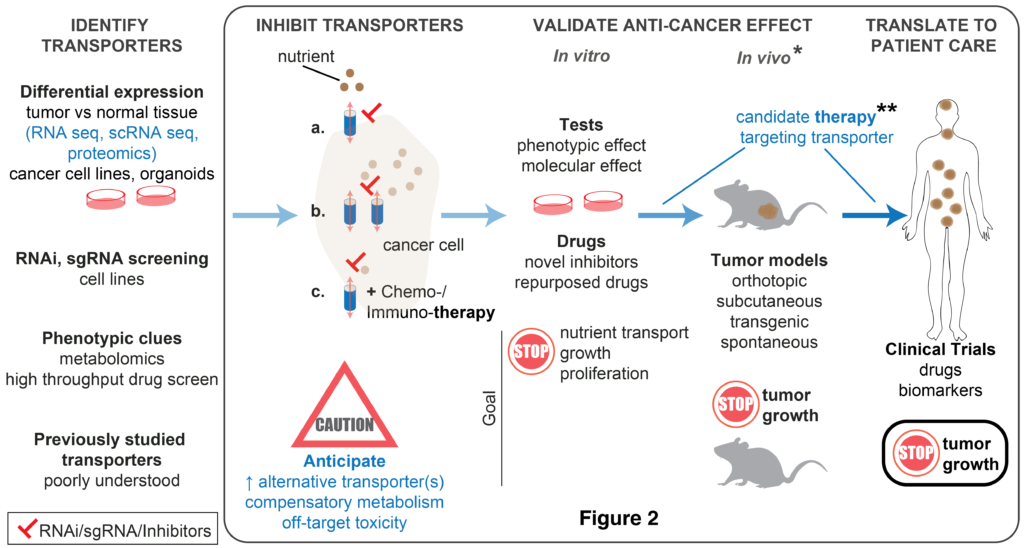
Targeting nutrient transporters
Membrane transporters are proteins that enable cells to obtain nutrients. Simply put, these transporters serve as the “mouth” through which cells “feed”. Studies have shown that in order to support growth, cancer cells hijack transporters for the purpose of excessively consuming nutrients such as glucose and amino acids. As depicted in Figure 2 (Source: Nwosu et al., 2023, Oncogene), various strategies may be adopted to identify and study transporters that enable cancer survival, growth, progression, metastasis or immune evasion. We are interested in identifying and molecularly characterizing transporters that are potential therapeutic targets in pancreatic cancer. This project can be pursued separately or under the GSH project.
Uncovering novel metabolic adaptation and drug resistance mechanisms
Drug resistance is a major barrier to cancer therapy. Finding ways to prevent resistance will improve the treatment of cancer patients and save lives. Metabolic adaptation is one mechanism by which cancer cells resist treatment. In essence, what the cancer cells do is to switch to using alternative nutrients when they are prevented from using their preferred nutrient. By so doing they can continue to survive and grow. For example, we have shown that although pancreatic cancer cells are highly reliant on glucose, they switch to using uridine when glucose is withdrawn (Nwosu et al., 2023, Nature, see Figure 3). Blocking the use of uridine via uridine phosphorylase (UPP1) suppressed tumor growth. We are interested in finding other bypass pathways and nutrients that support pancreatic cancer cells. Our hope is that blocking such adaptation could help overcome cancer drug resistance in patients.

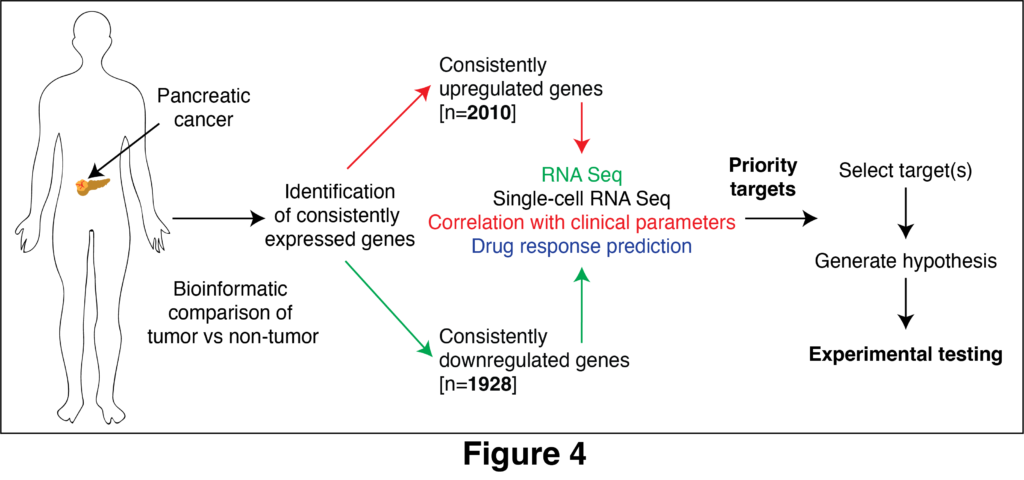
Characterization of priority targets
We have identified a number of genes that are often upregulated/high or downregulated/low in pancreatic cancer. We further called some of the genes “priority targets” based on their consistent correlation with variables associated with cancer aggressiveness, e.g., proliferation, metastasis and overall survival (Nwosu et al., 2025, JCI Insight, In press). Our hypothesis is that several of the priority genes are important drivers of pancreatic cancer or potential biomarkers. For example, UPP1 highlighted above is one of the priority targets. Those “targets” also include several other metabolic genes as well as transporters, transcriptional factors, epigenetic regulators and genes that encode signal transduction molecules. Thus, similar to UPP1, we will experimentally test the role of other priority targets and we strongly believe this project offers multiple opportunities for novel discoveries. Prospective lab members will be supported to study selected priority genes in the context of cancer metabolism (see Figure 4). This project is particularly ideal for graduate students and postdocs who wish to develop a long-term project that could be used for career transition grant applications.
METHODS
The methods to be used for the above projects include metabolomics/stable isotope tracing, extracellular flux analysis (Seahorse), genomics/sequencing technologies, epigenetics, proteomics, gene interference (e.g., CRISPR/Cas9, shRNA, siRNA), growth and other phenotypic assays, cell culture, mouse models (including transgenic, subcutaneous and orthotopic implantation), dietary studies in mice, microscopy, flow cytometry, histology and bioinformatics. Lab members will have multiple opportunities to learn how to use these methods to tackle scientific questions.
Our lab has several cutting-edge equipment required to facilitate our research while providing students the opportunity to learn useful skills that can set them up for success in academia or the industry. Our equipment include:
- Cell culture incubators (including hypoxia module)
- EVOS™ Imaging System (Microscope) with On-stage Incubator (including hypoxia module)
- QuantStudio 3 qPCR Machine
- Applied Biosystems Thermal Cyclers (for PCR, molecular cloning, etc)
- TECAN Spark® Multimode Microplate Reader (for drug screening, biochemical assays, etc)
- TECAN NanoQuant Plate for DNA/RNA and Protein Quantification
- Western Blotting and Transfer as well as Protein Electrophoresis modules
- Vacufuge Plus Concentrator (for metabolomics sample preparation)
- Tissue Lyser III (for homogenizing tissues of all kinds)
- Seahorse XF Pro 96 (for profiling cellular respiration and extracellular acidification)
- Agilent 1290 Infinity II Ultra-High Performance Liquid Chromatography (UHPLC) (for metabolomics)
- Agilent 6459D Triple Quadrupole Mass Spectrometer (for metabolomics)
In addition to the above, our lab members can conveniently assess additional state-of-the-art equipment and facilities via the Cornell Institute of Biotechnology located in the same building here our lab is housed.
FUNDING
The Nwosu Lab is currently supported by funds from the following sources:
- NIH/NCI K99/R00 Pathway to Independence Career Development Award.
- College of Agriculture and Life Sciences, Cornell University.
Many thanks to the funders!
